MethylDetect Kit For DNA Methylation Assessment
Product inquiry
Send us your inquiry to Assay Specific Control System and we will answer as soon as possible.
| For Research use only. Not for use in diagnostic procedures. Version 2 |
Protocol
For 200, 500 and 2500 reactions
Store at -20°C upon arrival
Date: December 2019, v2
1. General Information
The performance of the EpiMelt kit has been tested on the LightCycler® 480 High-Resolution Melting platform (product number: 05015278001) and LightCycler® 480 High-Resolution Melting Master (product number: 04909631001) PCR reagents in 96 well plates.
The use of this platform is recommended in combination with the EpiMelt kit.
| Other RT-PCR systems capable of performing high resolution melting (HRM) can be used following further optimization of the experimental conditions. |
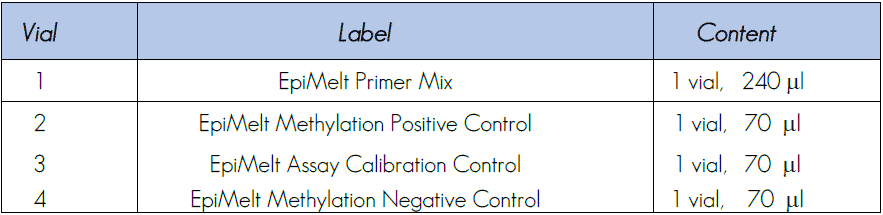
1.1 Contents:
1.2. Storage conditions:
The EpiMelt Kit is shipped at room-temperature.
When stored at -20C the reagents are stable through the expiration date printed on the label.
Avoid repetitive freezing and thawing of the kit components.
Shipping and temporary storage after opening for up to 8 weeks at 4°C has no detrimental effects on the assay performance.
For storage conditions of the LightCycler® 480 High-Resolution Master Mix, the associated protocol can be consulted for further details.
1.3. Additional equipment and reagents required:
- High-Resolution Master Mix (containing Taq polymerase, reaction buffer, dNTP mix, HRM dye, MgCl2, and water, PCR grade).
- An instrument combining Real-Time PCR with high-resolution melting capability.
- Plates or reaction tubes compatible with the Real-Time PCR and HRM instrument.
- Nuclease-free, aerosol-resistant pipette tips.
- Pipettes with sterile filter tips.
- Sterile reaction tubes for preparation of master mix.
Optional: Standard swinging-bucket centrifuge containing a rotor for multiwell plates.
1.2. Application
The EpiMelt Assay is designed and optimized for detection of DNA methylation status of the specific gene promoter region in human bisulfite converted genomic DNA. DNA, representing methylation positive, assay calibration, and methylation negative controls, are supplied with the assay to ensure straight forward data analysis and control optimal performance of the kit.
1.3. Preparation time
Assay time
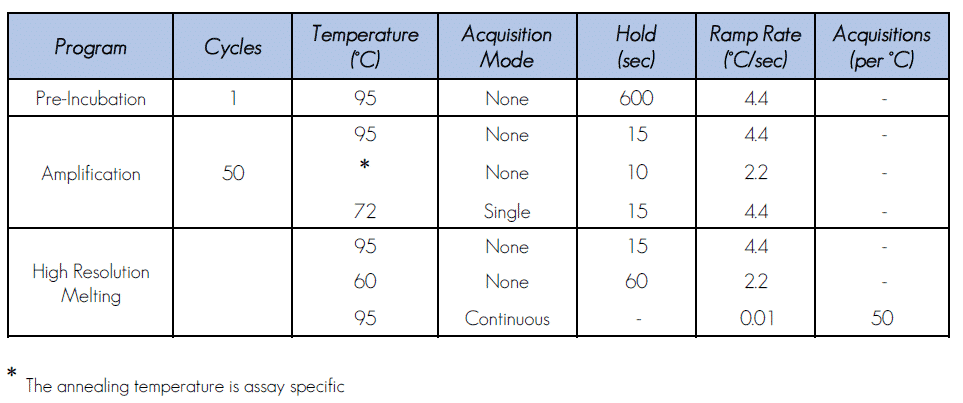
The cycling program for the EpiMelt Kit includes 10 min pre-incubation and 50 cycles of amplification followed by high-resolution melting and (approximately 90 -120 min when performed using the LightCycler®480 System).
2. Protocols
2.1. Before you begin
Sample materials
When using PCR for methylation studies, all DNA samples have to be bisulfite converted, to ensure preservation of the methylated cytosines in the template.
The changes induced by sodium bisulfite treatment in DNA are illustrated in figure 1:

Figure 1. The preserved methylated cytosines, which are part of a CpG dinucleotide, are underlined and in red. The unmethylated cytosines are converted to uracil, and further substituted with thymine during the PCR, and are illustrated in blue.
Bisulfite treatment converts unmethylated cytosines into uracil and leave the methylated cytosines unchanged. The DNA sequence of the methylated DNA strand is therefore different from the unmethylated. This results in different melting properties of the two PCR products, which are clearly distinguishable after HRM (figure 2).
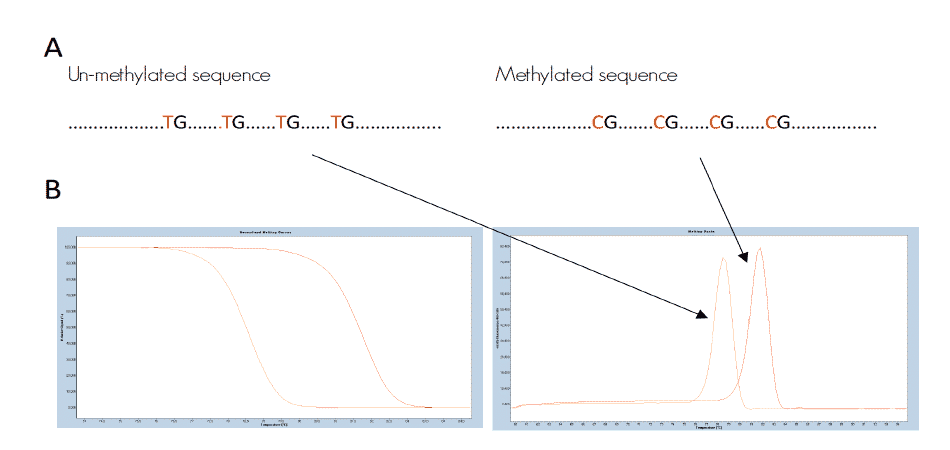
Figure 2. Schematic illustration of the principle behind HRM analysis. A) the difference in a DNA sequence after bisulfite conversion of a methylated and an unmethylated genomic region. B) the difference in melting properties of the PCR products from the methylated (red) and the unmethylated (orange) templates displayed as melting curves and melting peaks.
Sample DNA
Use 30 – 50 ng of bisulfite modified DNA per reaction. This is a theoretical concentration based on the DNA input for bisulfite conversion and the elution volume.
Kits for bisulfite conversion of the sample DNA can be purchased form other vendors.
! Follow the instructions from the bisulfite conversion kit for storage of the DNA after treatment.
The quality of the DNA should be suitable for PCR in terms of concentration, purity and absence of PCR inhibitors. Use the same DNA extraction procedure for all samples in the analysis.
To ensure sufficient quality of the DNA prior to bisulfite conversion analysis by a Bioanalyzer can be used to assess the DNA integrity, and Qubit Fluorometer is recommended for measuring the DNA concentration.
Assay calibration controls
A methylation positive, an assay calibration, and a methylation negative control are supplied with the EpiMelt Kit, and ready for use. The assay calibration control is included to ensure the assay sensitivity to detect methylation of1% in each experiment.
! The methylation positive, the assay calibration, and the methylation negative controls are ready to use and should not to be bisulfite converted.
The controls are each applied in duplicates or triplicates to all multiwell plates.
Negative Control Reaction
A No Template Control (NTC) should always be included in the analysis. The NTC contains the same reagents as the reactions for analyses, except that the DNA sample is replaced with the same amount of PCR grade water. The NTC should be present at each multiwell plate in triplicates.
Primers
A primer mix is supplied within the EpiMelt Kit, and the PCR assay conditions are given in Table 2.
2.2. Preparation of the PCR reaction
The protocol is calibrated using the LightCycler® 480 High Resolution Melting Master and the associated protocol can be consulted for further details 1.
The protocol is optimized using the LightCycler® 480 High-Resolution Melting Master (product number: 04909631001) PCR reagents.
Follow the procedure below in the given order to prepare one 20 ml standard reaction.
- Thaw the solutions and spin all tubes briefly in a micro-centrifuge before opening, to ensure that the content is collected at the bottom of the tube.
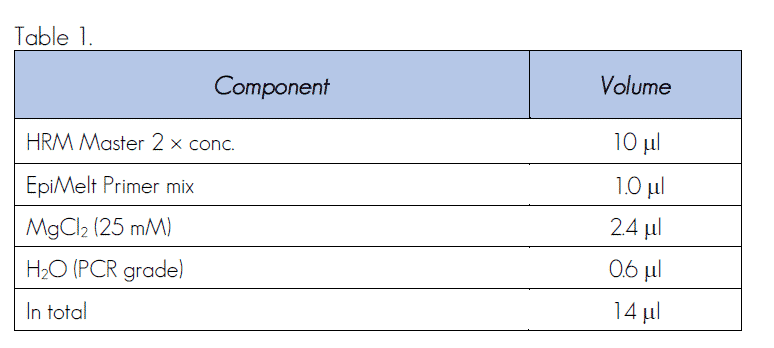
- Prepare the PCR mix for one 20 ml reaction by adding the following components in the order listed in Table 1 and keep it on ice.
Table 1
Multiple Reactions.
To prepare the PCR mix for multiple reactions, multiply the volume for each component with the number of reactions, plus adequate number of the reaction to accommodate provided in the kit control samples (methylation positive, assay calibration, and methylation negative control DNA) and the NTC .
! For example of the plate setup see figure 3.
Example:
To prepare a 96-well plate, in which all samples are analysed in triplicates including: 28 test (screened, unknown) samples (84 wells) + three different standards (9 wells) + NTC (3 wells) = 96 wells.
3. Mix the reagents carefully by pipetting up and down and spin briefly. Do not vortex
4. Pipette 14 ml PCR mix into 96 well plate.
5. Add 6 ml bisulfite treated DNA, corresponding to a theoretical calculated value of 30 – 50 ng DNA. Lower amount of template can be used, however, we recommend that the assay is optimized to the specific DNA concentration before processing the test samples.
! For each multiwell plate, add 6 ml of each standard control DNA provided in the kit (methylation positive, assay calibration, and methylation negative), preferably in triplicates.
6. Seal the multiwell plate with an appropriate sealing foil.
7. Spin for 2 min at 1000 ´
8. Place the multiwell plate in the instrument and start the PCR-HRM program.
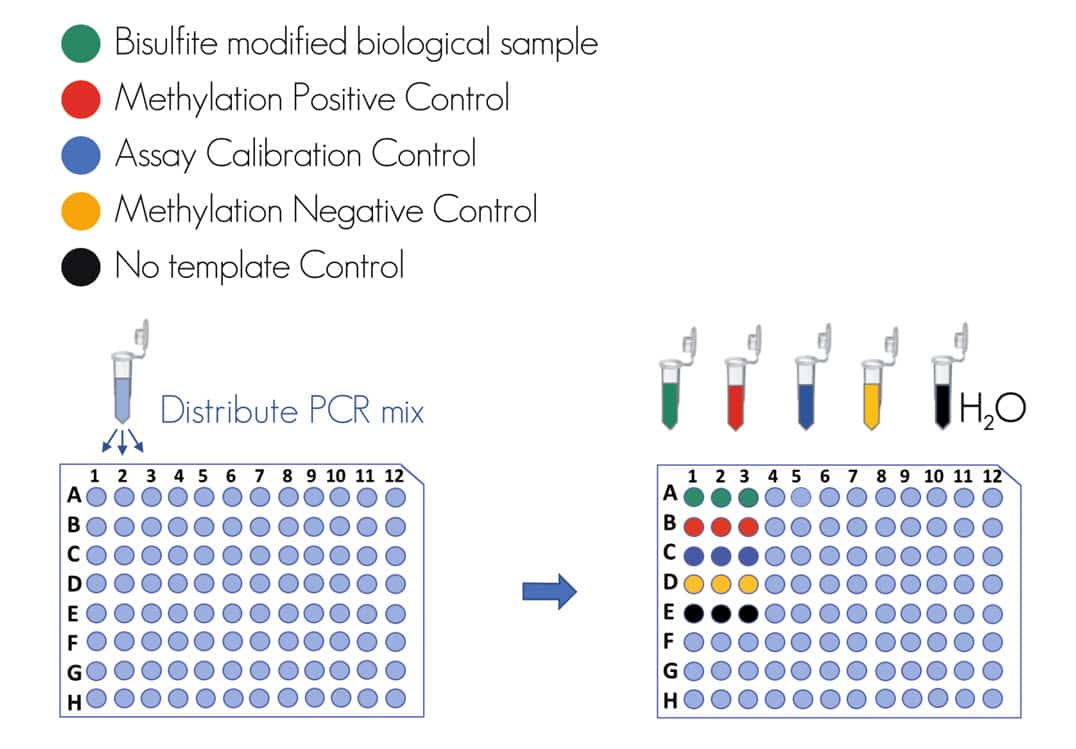
Figure 3
2.3. PCR and HRM program
The program below is suitable for the LightCycler® 480 System.
3. The principles behind the MS-HRM analysis method
Methylation-Sensitive High-Resolution Melting (MS-HRM) is a high-throughput technology for highly sensitive and in-tube analyses of locus specific methylation1,2.
The technology utilizes the difference in melting properties of the PCR product amplified from methylated and unmethylated DNA strands after bisulfite conversion (Figure 1).
The propraitory design of the MethlDetect MS-HRM assays makes them suitable not only to differentiate between methylated, un-methylated and mixes of these templates but also to identify heterogeneously methylated templates3 4 6.
4. Results
The results presented below were obtained using the EpiMelt Kit and the LightCycler® 480 High-Resolution Melting Master.
After the amplification part of the program, the PCR product, generated during the amplification, is subjected to high-resolution melting curve analysis, and the data analyzed using the LightCycler® Gene scanning or Tm calling Software.
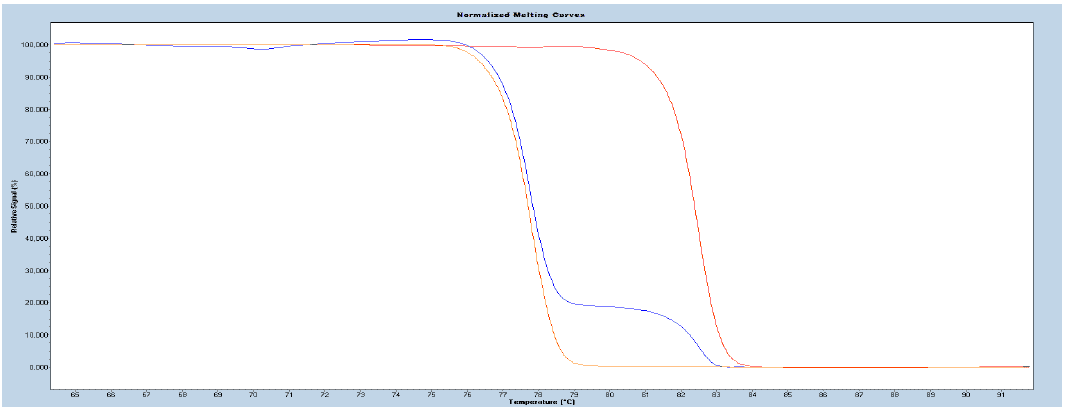
Figure 4. Normalized melting curves illustrating methylation positive control (red), the assay calibration control (blue), and methylation negative control (orange) of the gene specific templates supplied with the EpiMelt Kit.
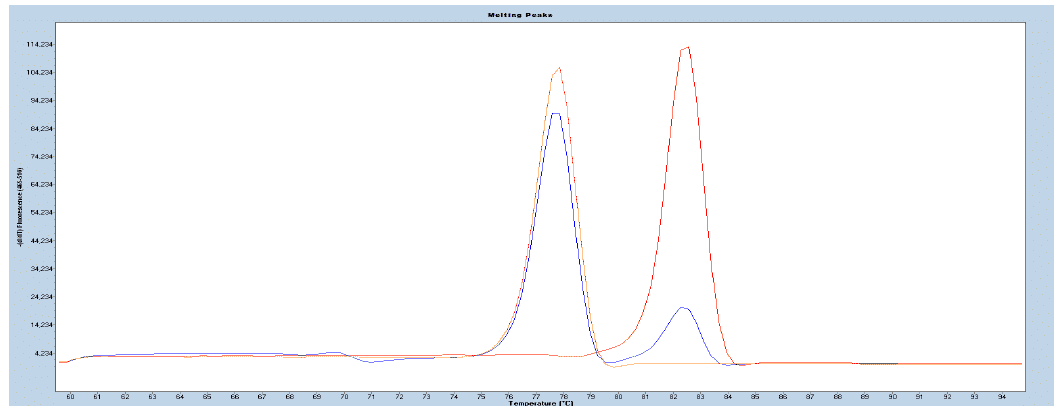
Figure 5. Relative signal difference (d/dT) plot illustrating the methylation positive control (red), the assay calibration control (blue), and methylation negative control (orange) of the gene specific templates supplied with the EpiMelt Kit.
References
- Clark SJ, Harrison J, Paul CL, Frommer M. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 1994;22(15):2990-2997.
- Wojdacz TK, Dobrovic A, Hansen LL. Methylation-sensitive high-resolution melting. Nat Protoc. 2008;3(12):1903-1908.
- Wojdacz TK, Dobrovic A. Methylation-sensitive high resolution melting (MS-HRM): a new approach for sensitive and high-throughput assessment of methylation. Nucleic Acids Res. 2007;35(6):e41.
- Wojdacz TK, Windelov JA, Thestrup BB, Damsgaard TE, Overgaard J, Hansen LL. Heterogeneous and low-level methylation of novel biomarker candidates for breast cancer clinical management. Cancer Research. 2014;74(19).
- Bock C, Halbritter F, Carmona FJ, et al. Quantitative comparison of DNA methylation assays for biomarker development and clinical applications. Nature Biotechnology. 2016;34(7):726-+.
- Hussmann, D. Hansen, L.L. Methylation-Sensitive High Resolution Melting (MS-HRM). Methods Mol Biol. 2018;1708:551-571.
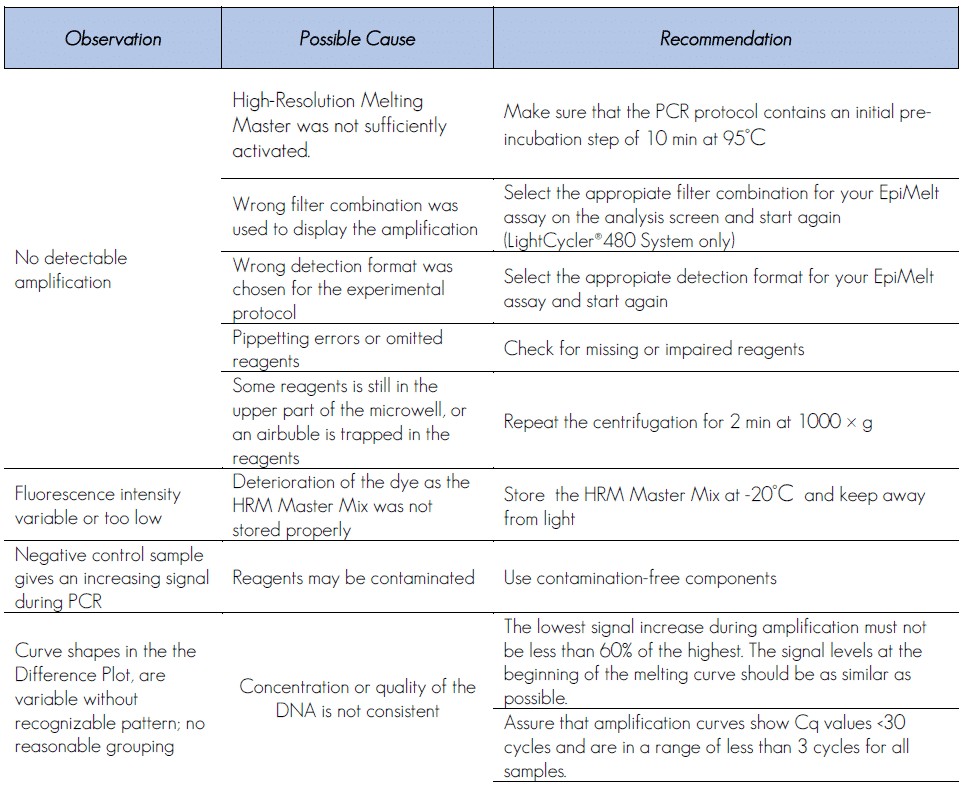
5. Troubleshooting
6. Ordering Information
MethylDetect offers a number of EpiMelt DNA methylation detection kits targeting specific genomic regions. For a complete overview of the products and ordering information please visit www.MethylDetect.com
7. Supplementary Information
License Disclaimer
For patent licence limitations for individual products please refer to www.MethylDetect.com
Regulatory Disclaimer
For Life Science research only. Not for use in diagnostic procedures.
Safety Data Sheet
Please follow the instructions in the Safety Data Sheet (SDS) at www.MethylDetect.com
Contact and Support: www.MethylDetect.com
©2022 MethylDetect, all rights reserved
| For Research use only. Not for use in diagnostic procedures. Version 2 |
